
Designed by Freepik
Genomes4Health (G4H) is led by the Weimer lab at UC Davis. It was initiated with whole genome sequencing (WGS) in 2012 as a landmark global consortium (The 100K Pathogen Genome Project) that sought to use NGS to examine bacterial genomes in public health for the identification of outbreaks and inclusion of isolates within a disease setting. Today, this work has expanded beyond WGS to encompass metagenomics in food, disease, and agriculture – much more than was envisioned with WGS alone. Using advanced sequencing approaches we have uncovered an unexpected vastness of diversity of bacterial genotypes that form the basis of identification and tracking using population genomic approaches that are only recently available to microbiology. Continual bacterial genome evolution is hindering our ability to consistently detect and mitigate pathogens, which interfere with our preparedness to defend public health. By leveraging genome diversity, it enables new biotechnology, diagnostic, and public health approaches for the management of diagnostics and phylogeny.
The expansion for metagenomes, using metaRNAseq, is informed by bacterial genome sequence and the population diversity to enable analysis of population genomics in bacteria using machine learning and bacterial metabolism. Bacterial identification, microbiome impact on health, functional biomarker discovery, source tracking in agriculture, public health outbreaks, bacterial survival, virulence, and the environmental modulation of metabolism is fully represented in projects and available sequences. Of critical importance within G4H is of bacterial persistence and microbiome assemblage in disease settings for humans and animals. A goal of G4H is to understand the link between disease, the microbiome, and bacterial evolution on a global scale by using population genomics concepts based on WGS and metagenomes.
Use of population genetics tools and concepts is a bold and revolutionary approach that is new to microbiology at this scale. Genomics is empowering precise and robust molecular testing that can be used as a forensic tool for many applications across humanity. In spite of extensive efforts to increase regulation and develop early warning diagnostics for improved public health monitoring, definitive interventions remain elusive in large, partly due to the lack of sufficient information about microbial diversity. This effort is aimed at linking diagnostics with functional characteristics encoded in the genome to enable persistence in the environment, mediate zoonotic transmission, and allow host adaptation. G4H collaborators are focused on tackling these global problems using big data approaches that provide answers from genotype to allelic variation that are coupled with disease and persistence. This work ushers in a new era of bacterial study – microbial population genomics.
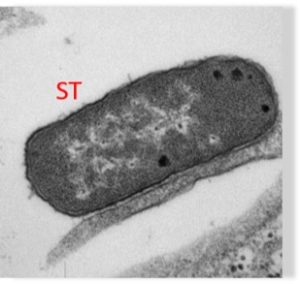
Salmonella infecting a human stem cell Weimer Lab
Bart C. Weimer Ph.D., Professor, UC Davis, School of Veterinary Medicine
1089 Veterinary Medicine Drive, Davis, CA 95616 (530) 752-6426
bcweimerATucdavis.edu


